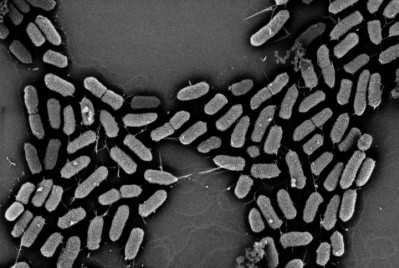
E.coli research targets quicker quantification and detection

The test is a molecular assay, or polymerase chain reaction (PCR), that detects bacteria based on genetic sequences, or the bacteria's ‘fingerprints’.
It can be used in a diagnostic or research laboratory to detect the pathogen and help with quality control in cattle facilities.
The researchers said it could help the cattle industry save millions of dollars each year by earlier and accurate detection of E. coli.
Project validation
Lance Noll, master's student in veterinary biomedical science, Greensburg; T.G. Nagaraja, professor of diagnostic medicine and pathobiology; and Jianfa Bai, assistant professor in the Kansas State Veterinary Diagnostic Laboratory are leading the project.
Noll said the method has only been validated for feces but he was confident that it can be used in other areas, such as carcass samples.
“Although a number of methods are available, only few have been developed for quantification of the organism,” he told FoodQualityNews.com.
“The quantification allows us to identify those cattle, called ‘super shedders’, and is strong evidence that super shedders are a major source of spread among animals in a herd and carcass contamination in abattoirs.”
The novelty of the test is that it targetsfour genes, said the researchers.
“All the previous Real Time-PCR methods for O157 involved three or fewer genes. A very limited number of diagnostic microbial assays have been successfully developed that are truly able to co-amplify more than three target genes.”
Centers for Disease Control and Prevention figures show that STEC causes 230,000 cases of illness a year and slightly more than 1% results in hospitalization and life-threatening complications.
Culture-based and PCR
Nagaraja said culture-based methods are time-consuming and labor intensive and PCR has the advantage of being rapid and high-throughput plus being amenable to automation.
“Once the samples are ready, it takes about two hours to run the assay and we can do up to 96 samples in two to three hours.”
Two more assays have been developed as part of the research, said Nagaraja.
“We also have developed two more assays to cover the six major groups of non-O157 STEC (O26, O45, O103, O111, O121, and O145). Another graduate student, Pragathi Shridhar, is working on this.
“Our strategy is to use three sets of PCR assays to detect seven serogroups and three virulence genes in fecal samples.”
The researchers are part of a College of Veterinary Medicine team studying preharvest food safety in beef cattle.
Kansas State University is one of 15 universities involved in the STEC research that was funded by a five-year, $25m grant from the US Department of Agriculture.
Other objectives are detection, biology, risk analysis and assessment, risk management and communication and involve researchers from the University of Arkansas, California-Davis, California-Tulare, Delaware, New Mexico State, North Carolina State, Texas A&M and Virginia Tech.